Our Research
Advancing the field of computational pathology through innovative AI solutions
Research Projects
Our multidisciplinary approach to solving complex problems
Multimodal AI for Breast Cancer Subtype Analysis and Precision Therapy Recommendation
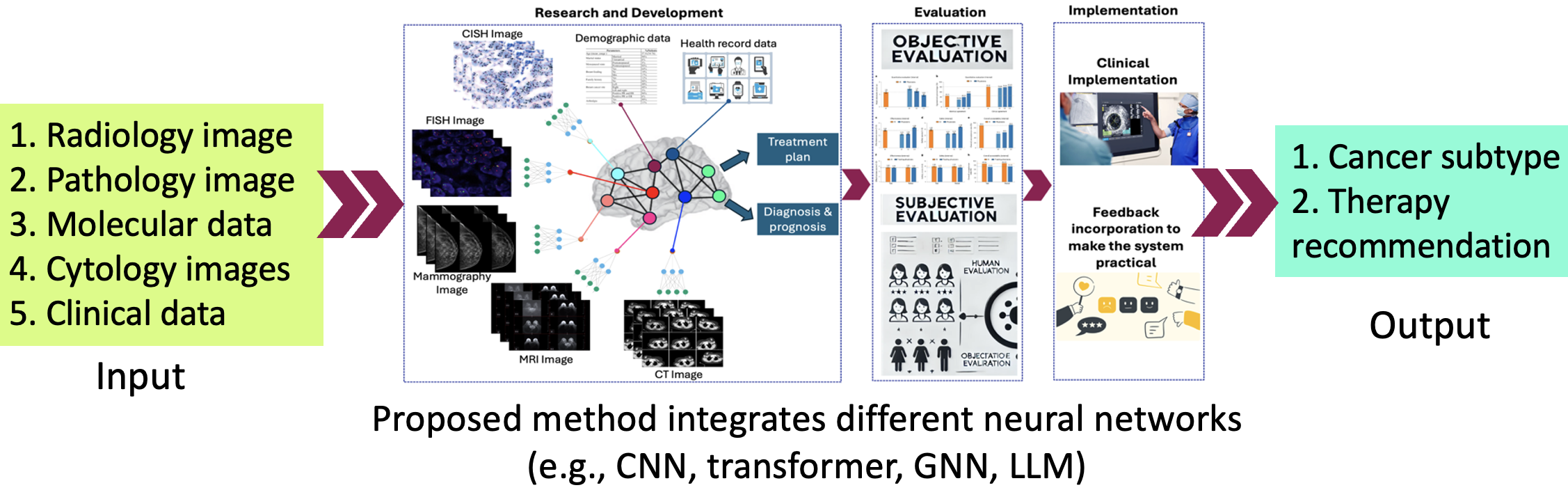
This project aims to develop a multimodal AI system that analyzes diverse breast cancer data—including histopathology, clinical, genetic, and imaging data—to accurately determine cancer subtypes and recommend personalized treatment. The system is designed to improve diagnostic precision and reduce reliance on expensive, expert-driven methods. By integrating all relevant data sources, the AI model will provide a low-cost, scalable solution that supports precision medicine and expands access to high-quality breast cancer care.
Automated Diganosis Pipeline for Breast Cancer Metastasis Assessment
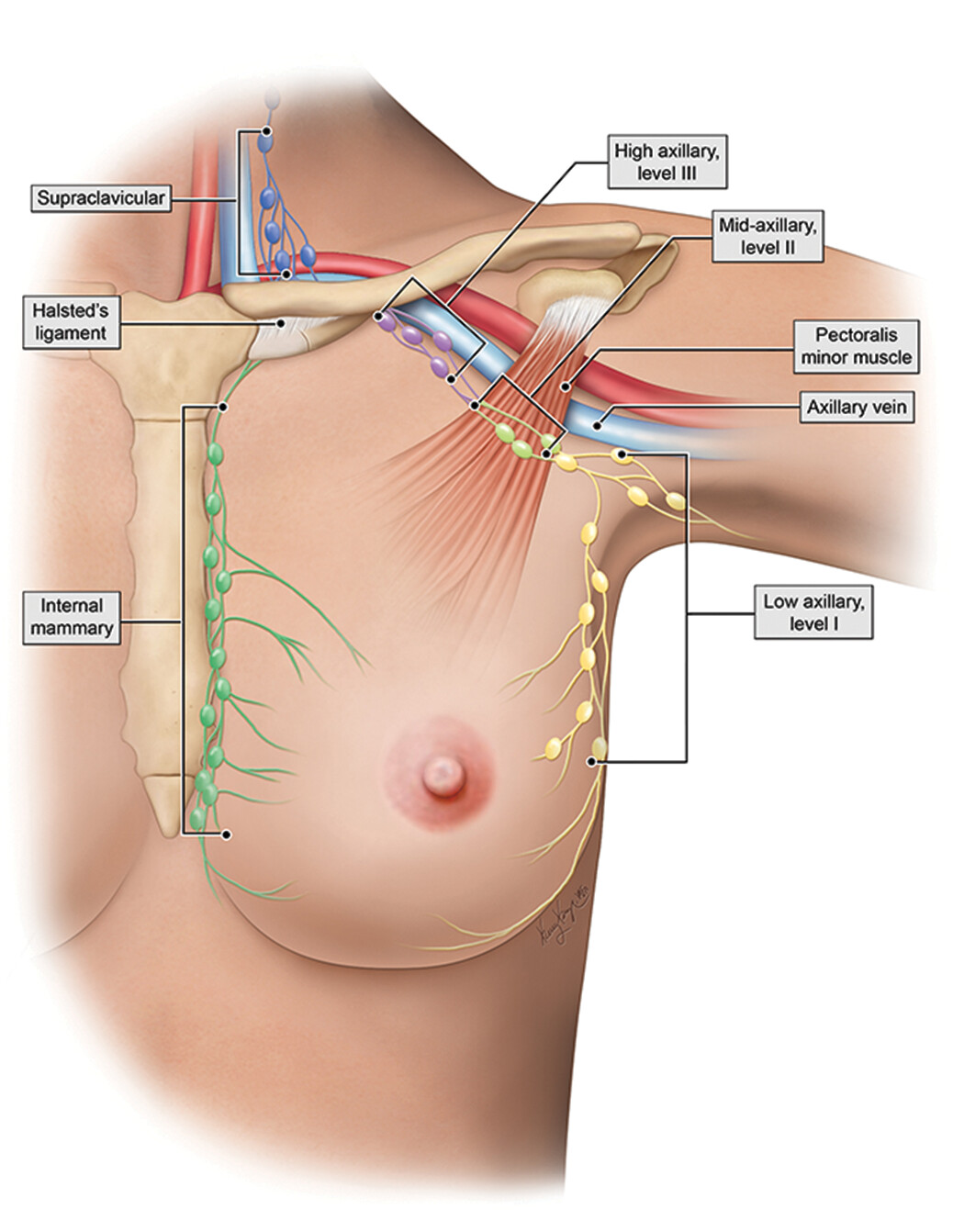
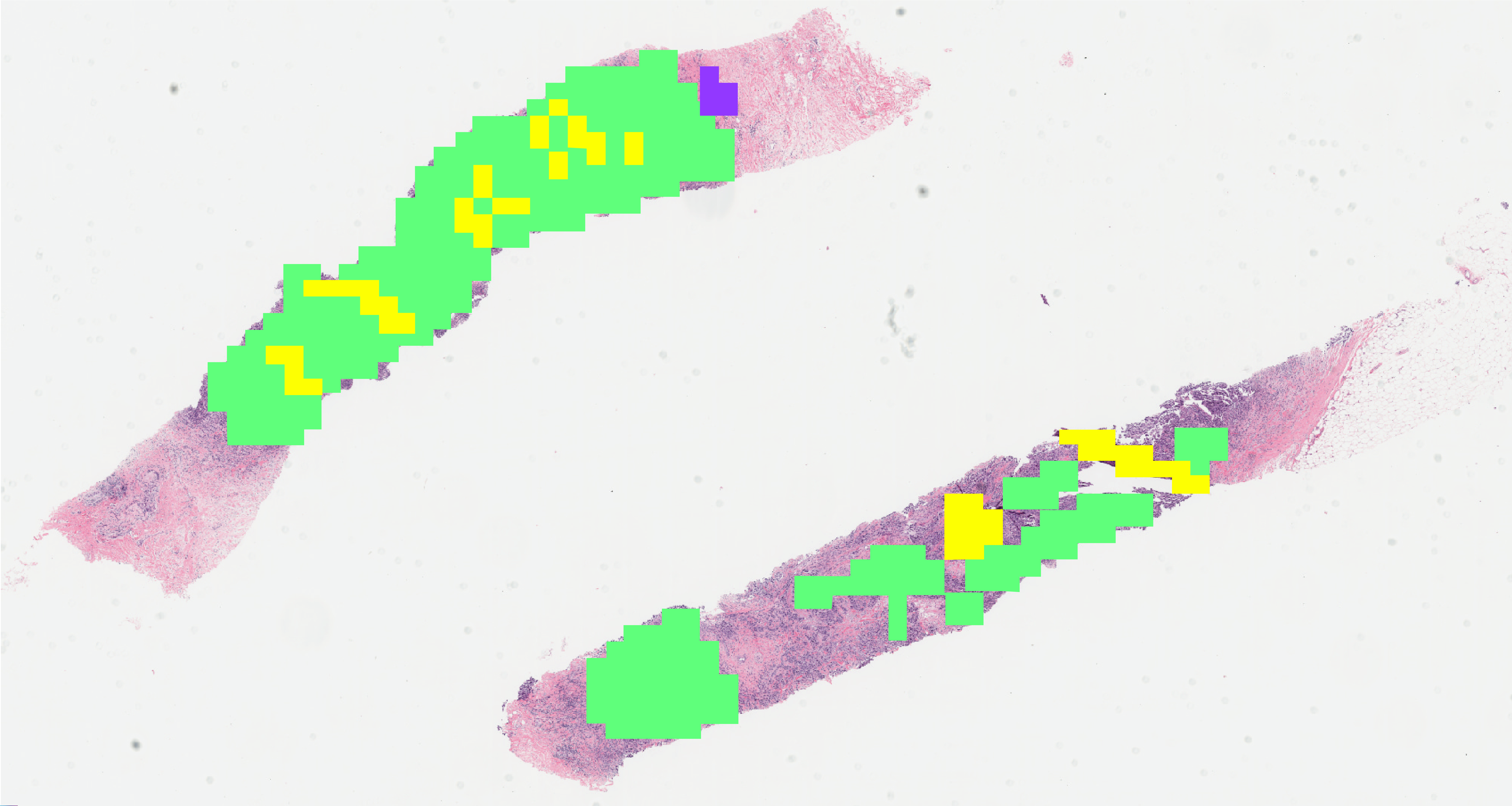
This research aims to develop an AI-guided automated pipeline for assessing breast cancer metastases in distant and axillary lymph nodes using affordable digital images of H&E-stained biopsy specimens. Traditional imaging methods (e.g., MRI, CT, PET-CT) often fail to detect early metastases and depend heavily on expert interpretation. While biopsy-based assessment is more accurate, it is underutilized in low-resource settings like Bangladesh and other developing countries due to its labor-intensive nature. Our system captures biopsy images using a camera-mounted microscope, identifies tumor regions using a transformer model, and classifies metastases using a graph neural network (GNN). The pipeline delivers fast, accurate, and cost-effective results, helping pathologists make informed decisions. This research directly supports the UN Sustainable Development Goals (SDGs), including Good Health and Well-being (SDG 3), Industry and Innovation (SDG 9), Reduced Inequalities (SDG 10), and Gender Equality (SDG 5).
HER2 Grading of Breast Cancer Patients
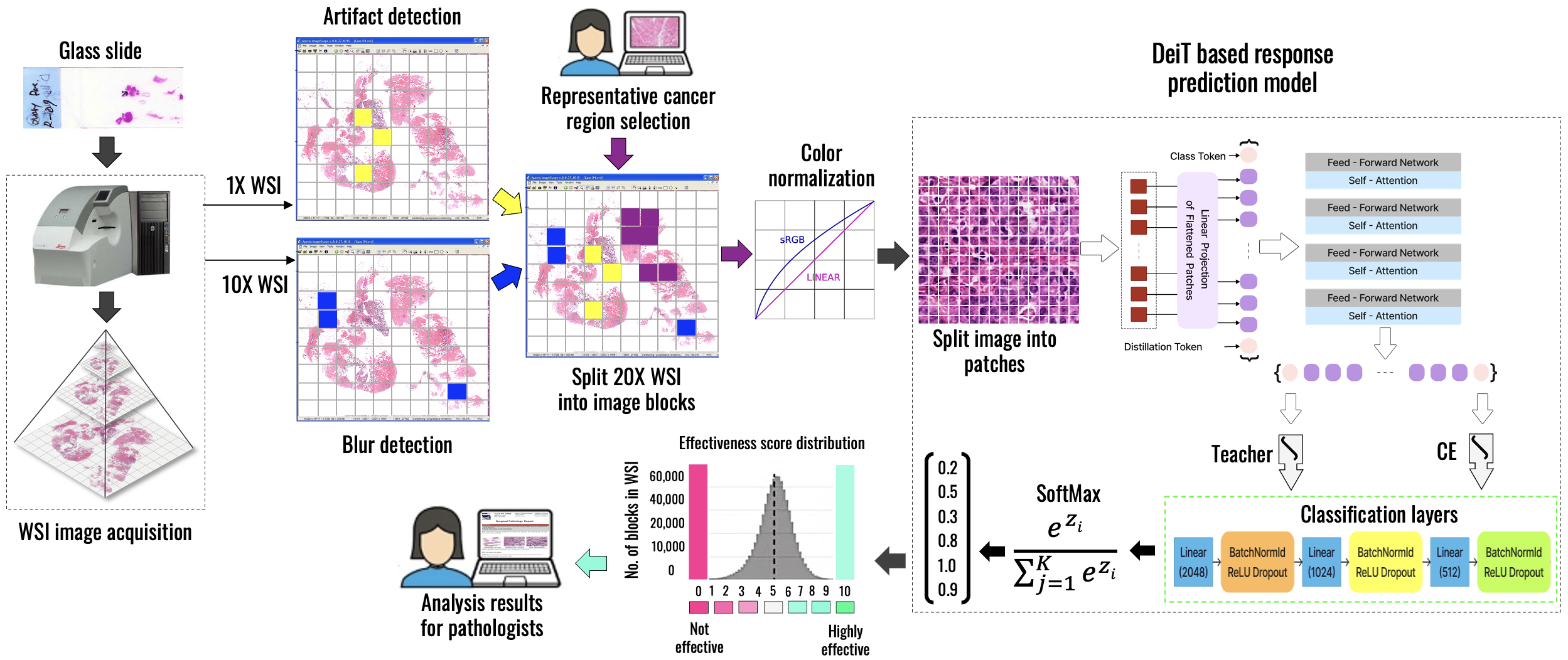
Residual Tumor Cellularity Assessment After Neoadjuvant Therapy
Residual Tumor Cellularity (RTC) assessment after neoadjuvant therapy is essential for evaluating treatment response and guiding further clinical decisions in breast cancer care. We intend to develop a comprehensive and practical system to assess tumor cellularity from H&E-stained whole slide images. This project intends to leverage multimodal AI to support precise prognostic evaluation, enable timely treatment adjustments, and facilitate large-scale, standardized pathology analysis in both high- and low-resource settings.
AI-guided Solution for Selecting Ovarian Cancer Patients for Bevacizumab Monoclonal Therapy
Ovarian cancer treatment outcomes can vary significantly among patients, making personalized therapy selection crucial. This research aims to develop an AI-guided solution leveraging advanced multimodal artificial intelligence techniques to predict which ovarian cancer patients will benefit from Bevacizumab monoclonal therapy. The multimodal AI must provide accurate, rapid, and reliable predictions by analyzing diverse data sources—including histopathological whole slide images, clinical records, and genomic information. This solution is intended to enhance treatment effectiveness and help reduce unnecessary side effects and healthcare costs.
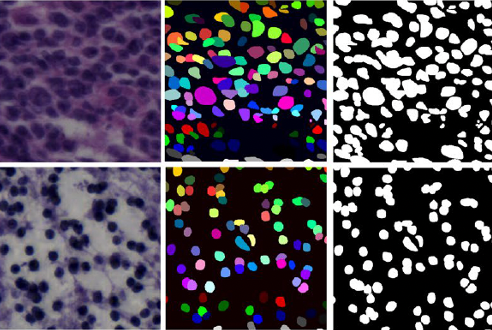
Nuclei Segmentation in Histopathology Images
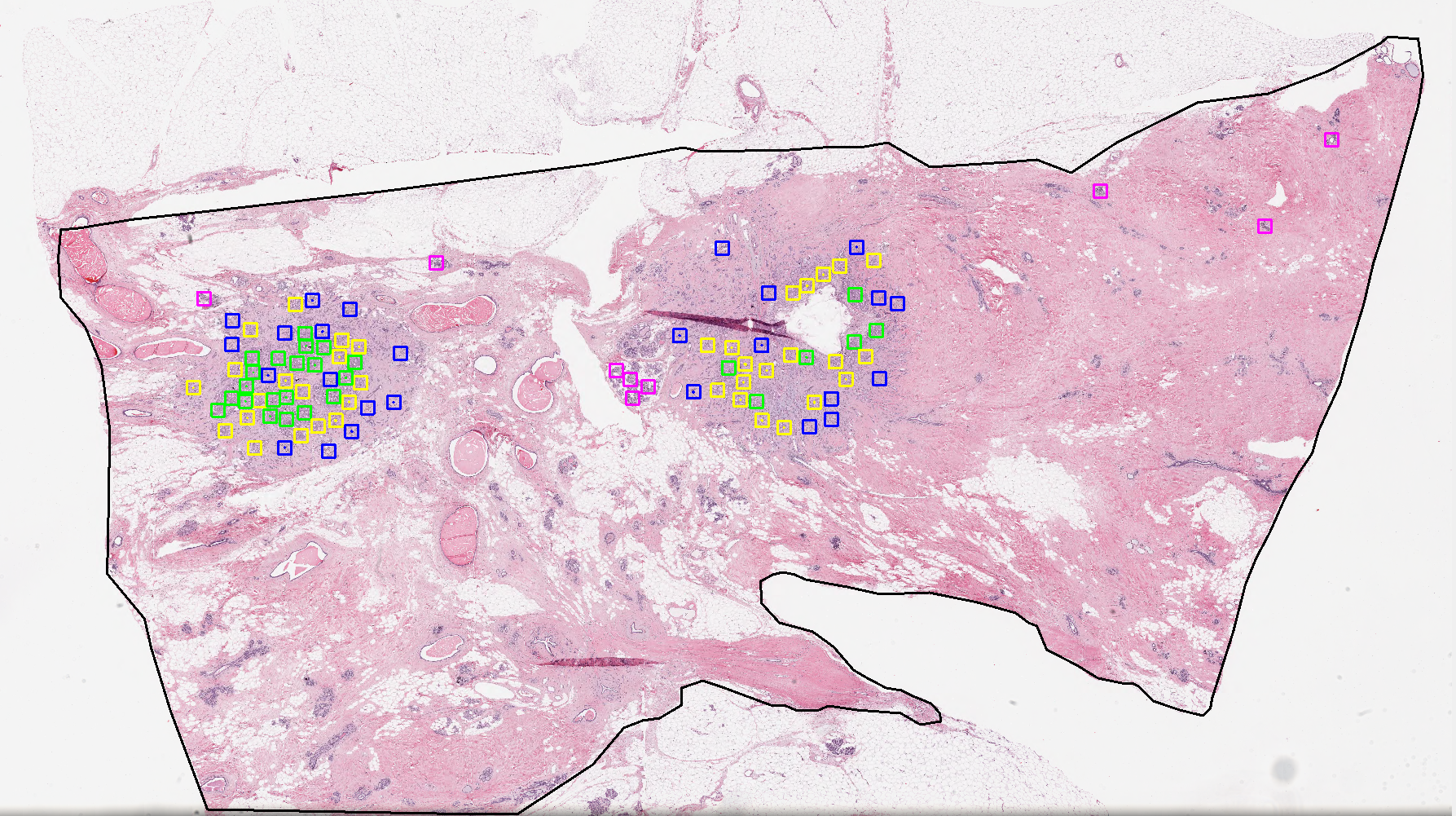
This research project focuses on developing advanced AI algorithms for accurate nuclei segmentation in histopathology images, a critical step in computational pathology. Precise segmentation of cell nuclei is essential for downstream tasks such as tumor grading, biomarker quantification, and patient prognosis. The project leverages deep learning models, including convolutional neural networks (CNNs) and transformer-based architectures, to detect and delineate nuclei across diverse staining conditions and tissue types. By addressing challenges such as overlapping nuclei, staining variability, and complex tissue morphology, this work aims to improve the reliability and scalability of digital pathology workflows and support more precise and automated cancer diagnosis.
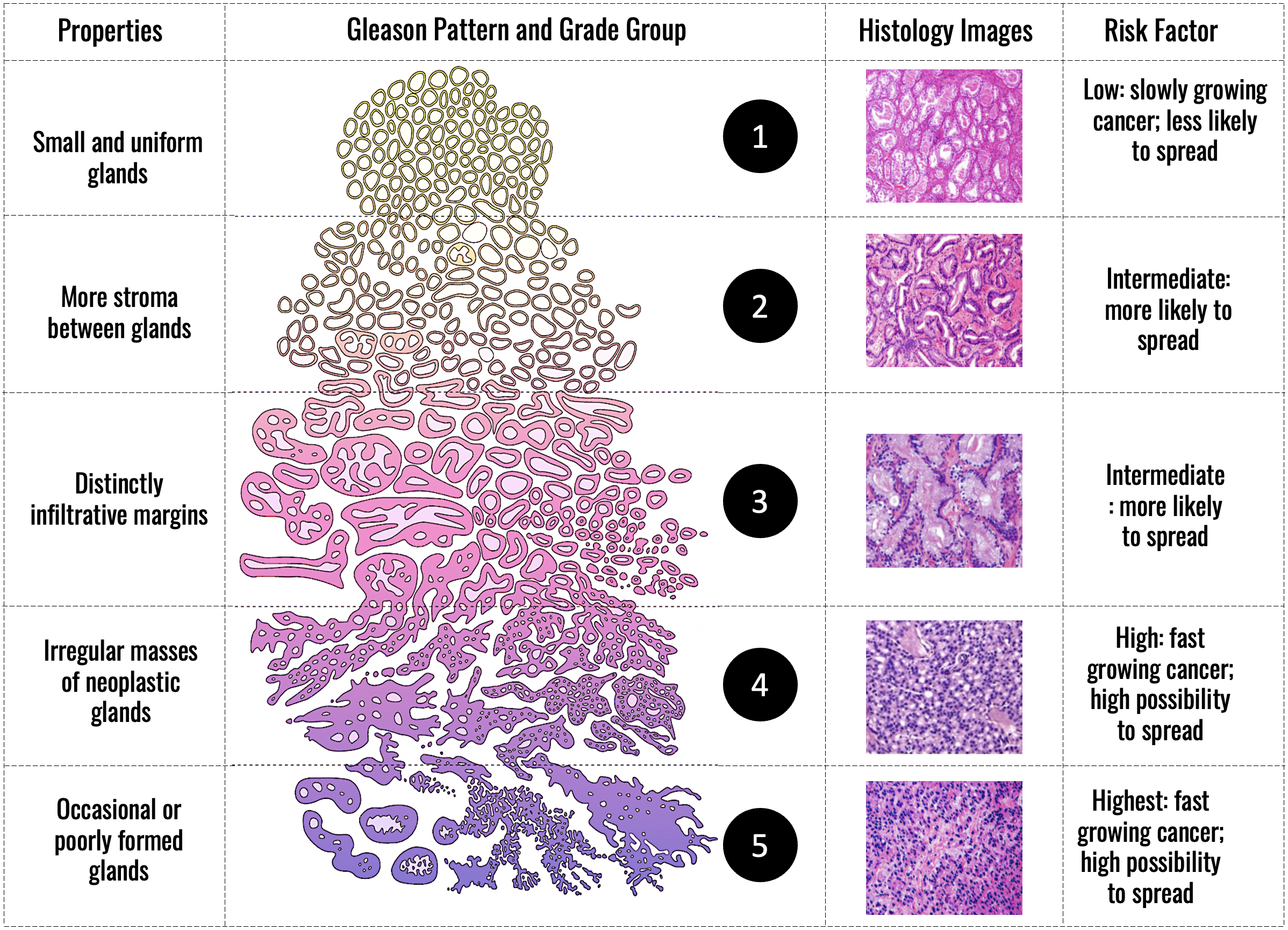
Gleason Grading of Prostate Cancer
This research focuses on developing deep learning-based methods for automated Gleason grading of prostate cancer using low-resolution histopathology whole slide images. Gleason grading is a critical step in assessing tumor aggressiveness and guiding treatment decisions. However, manual grading is time-consuming and subject to inter-observer variability. By leveraging advanced deep learning techniques, this project aims to provide a fast, consistent, and scalable solution that supports pathologists in clinical decision-making—even when only low-resolution images are available.
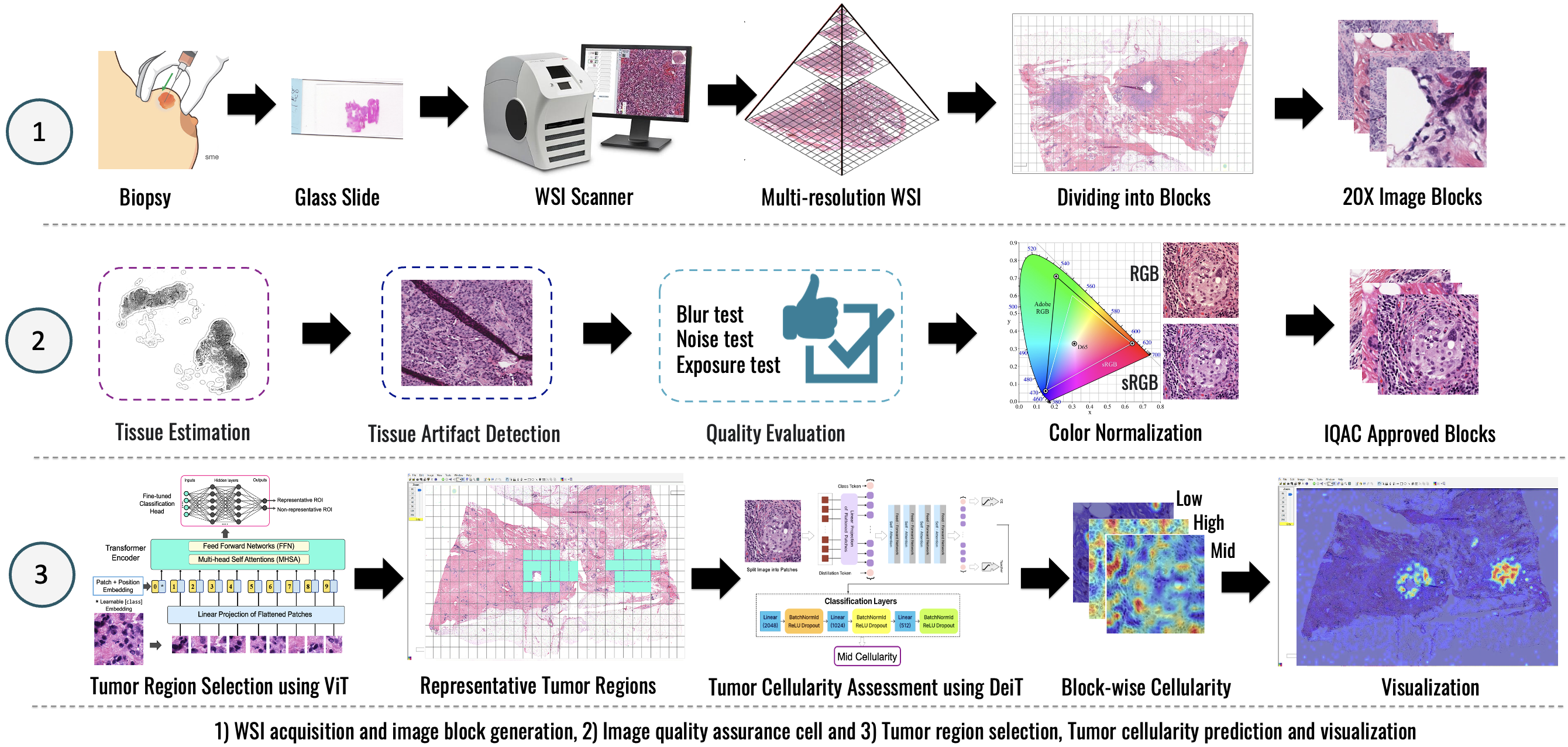
Quality Evaluation for AI-assisted Medical Image Analysis
This research focuses on advancing image quality evaluation techniques to enhance the performance and reliability of AI-assisted medical image analysis across diverse imaging modalities. This includes whole slide images (WSIs) in digital pathology, endoscopy images, and other clinical imaging data where quality variations can significantly impact diagnostic outcomes. This project aims to develop more robust AI models, improve data preprocessing pipelines, and contribute to more accurate and dependable clinical decision-making by systematically assessing issues such as blurriness, artifacts, noise, and color inconsistencies.
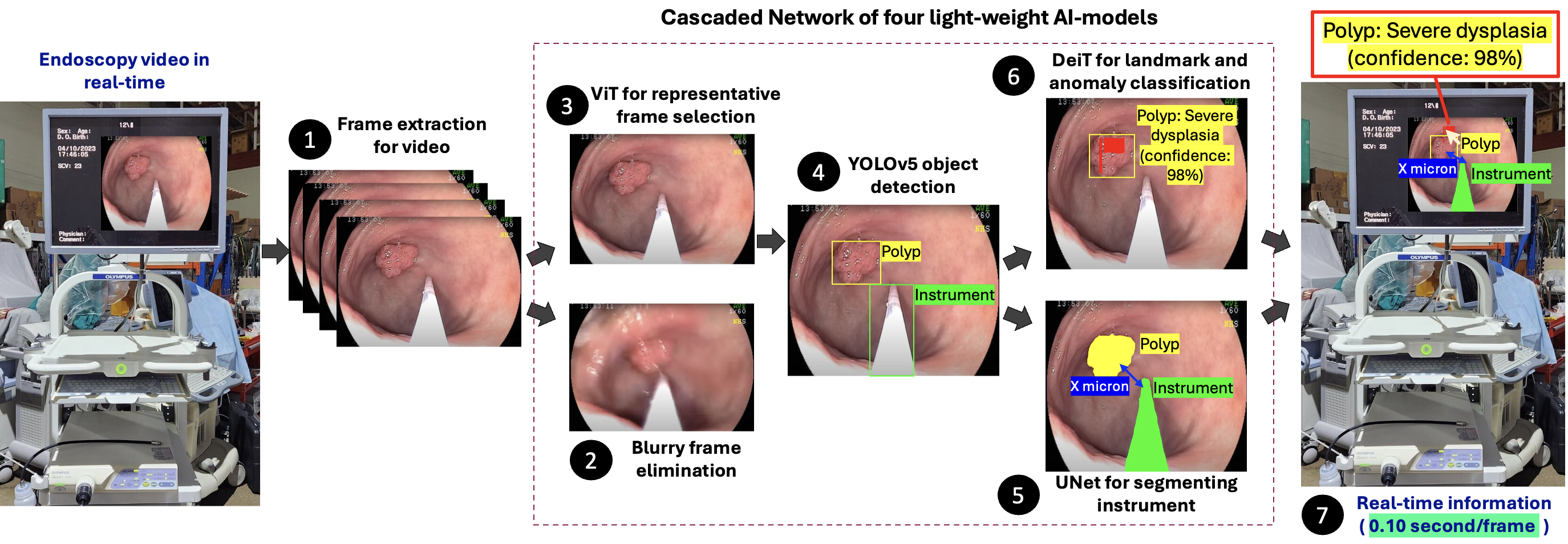
Endoscope vision enhancing ease for endoscopists and comfort for patients
This project aims to develop an AI-guided vision module for endoscopes to assist in real-time navigation, anomaly detection, and surgical procedures within the digestive tract. Traditional endoscopy is labor-intensive and prone to patient discomfort, diagnostic errors, and endoscopist fatigue. Our system uses artificial intelligence to identify landmarks, anomalies, and surgical tools during endoscopy, helping guide smoother and safer procedures. It improves diagnosis accuracy, reduces injury risk, and supports endoscopists with real-time insights. This research will be conducted in collaboration with medical institutions in Bangladesh and will build on our previous study in polyp detection and anatomical landmark recognition.
Computer Vision
This project develops computer vision–based AI solutions that use image data and information technology to automate complex tasks, improve accuracy, and support efficient decision-making across various real-world applications.